# Statistical genetics software
WARNING
This sofware is no longer actively maintained
# CoNVEM

Copy Number Variation Expectation Maximisation program which graphically and numerically presents CNV allele and genotype distributions. Complex CNVs (eg the salivary amylase locus) are typically assayed as a total copy number (aggregating both allelic copy numbers). CoNVEM can be used to determine the most likely population allele and genotype frequencies from these copy number counts, enabling inferences to be made about mutational history and population allellic architecture. Hum Mutat. 2010 Jan 14.
# CubeX

CubeX provides the exact solutions to Hill's cubic equation for haplotype frequency between two loci. Whilst this is often satisfactorily solved using an expectation maximisation approach, under certain circumstances more than one biologically possible solution to the equation may exist. Given the nine diplotype counts for a pair of biallelic markers, CubeX will present all biologically possible solutions. BMC Bioinformatics 2007, 8:428.
Code available on GitHub
# Hardy-Weinberg equilibrium calculator

An HWE calculator with analysis for ascertainment bias. This program is particularly relevant to Mendelian Randomisation analyses, where it is important to evaluate missingness under different genetic models. Analagous to the "intention to treat" analysis in RCTs, MR analyses should include "intention to analyse". The calculator is also of general utility as a quick test of Hardy-Weinberg equilibrium. Am J Epidemiol. 2009 February 15; 169(4): 505514.
# MIDAS
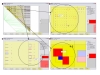
MIDAS implements an expectation-maximisation approach to the analysis of interallelic linkage disequilibrium analysis between any pairwise combination of multiallelic and biallelic markers. An interactive graphical interface enables the user to visualise the linkage disequilibrium across many markers. BMC Bioinformatics 2006, 7:227